 |
Breeding Value Estimation for Animal Breeding Programs |
MiXBLUP 2.2Breeding value estimation for Animal Breeding Programs using pedigree and genomic informationProduct informationMiXBLUP is a modern genetic evaluation system for all breeding organizations. It estimates genetic merit of individuals using observations and genetic similarity, given the components of variance. MiXBLUP offers the potential to accelerate genetic progress in your breeding population. MiXBLUP is easy to use, fast, suitable for simultaneous analysis of a large number of traits and able to fit complex models.It supports a wide range of methods to specify genetic similarity between individuals, such as pedigree relationships, genomic relationships, and regression on SNP covariates. 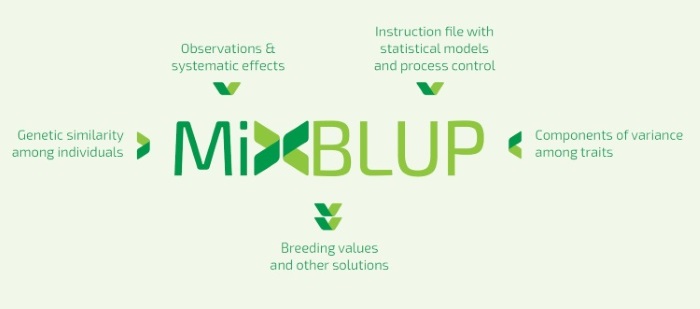
AvailabilityThe MiXBLUP software can be downloaded from this website. MiXBLUP only works with a license, which can be ordered on this website.A 30-days free trial license is available for evaluation purposes. A trial license allows you to do a full run, regardless of the number of equations, to be able to evaluate its performance, but the output contains only the first 1,000 solutions. A commercial license is available for routine use. A small license at reduced cost is available for breeders with a relatively small population and pedigree relationships only. For large evaluations or the use of genomic relationships, a full license is required. AdvantagesMiXBLUP allows you to use sophisticated models in estimation of breeding values for all animal species. MiXBLUP uses a fast, state-of-the-art solving algorithm. This ensures fast computation of estimated breeding values even if your evaluation model is complex and your animal population is large. MiXBLUP can be used in practice for modern applications, such as random regression models and social interaction models. Genetic similarity between individuals can be specified with pedigree and genomic information. Gene and marker assisted genetic evaluation is also supported.OriginMiXBLUP is developed by scientists of LUKE National Resources Institute Finland and the Animal Breeding & Genomics Centre of Wageningen UR Livestock Research in the Netherlands. The core of MiXBLUP is derived from the MiX99 software, developed by LUKE National Resources Institute Finland.MiXBLUP is currently used for routine evaluation of breeding populations of dairy cattle, beef cattle, pigs, layers, dairy goats, sheep and dogs. SupportThe user license includes support for installing the software and for software related problems. Extensive documentation on MiXBLUP is available on this site. For an additional service fee also individual support is available for issues related to setting up models.Features MiXBLUP
Output MiXBLUP
Possibilities MiXBLUP
|